
 |

|
Some people compare the complementary strands of DNA and the way they come together to the way a zipper flows together and locks its teeth. It’s not a bad analogy, and it can teach us something about the information value of the DNA code.
Consider the zipper itself as a kind of “one-letter DNA.” Each tooth on either side of the opening is identical, with a bump on one surface and a bump-sized depression or hole on the other.1 The slider as it moves upward aligns the teeth and meshes them, so that the bump on a tooth on this side fits into the hole on the back of the tooth ahead of it on that side. Lateral pressure keeps the two locked together. If we tried to read the zipper’s teeth as a kind of code, like DNA, the message would be very boring: “dit-dit-dit” on one side, “dot-dot-dot” on the other. It would have no information value. It would not even be a nonsense code but a no-sense code, useless except maybe for counting the teeth.
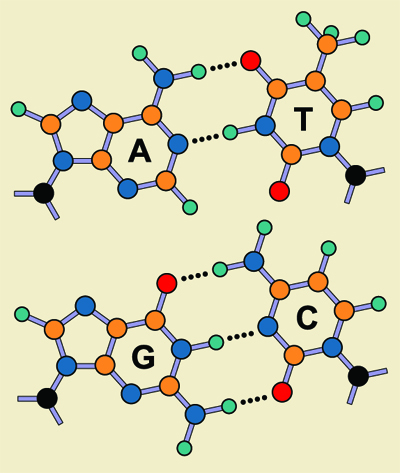
DNA, on the contrary, has a rich information value because it contains four kinds of teeth. The backbone of the zipper—the webbing band into which the teeth are sown or fused—is a series of ribose sugar rings, containing one oxygen and five carbons. They are connected up and down the zipper by phosphate groups that attach the fifth carbon on one ribose ring to the third carbon on the next ring along the strand. The first carbon on each ring is where the working “teeth” are attached, well away from the webbed backbone. Those teeth are made of more ringlike molecular structures called purines and pyrimidines.
Two of the teeth—the bases adenosine, or A, and guanine, or G—have a nine-member, double-ring structure that contains four nitrogen atoms and five carbon atoms, called a purine. The other two teeth—the bases cytosine, or C, and thymine, or T2—have a six member, single-ring structure containing two nitrogens and four carbons, called a pyrimidine. One of each of these pairs—C from the pyrimidines, and G from the purines—has three attachment points available for covalent bonding, or the sharing of electrons between nearby atoms in a molecule. The other of each pair—A and T—has only two attachment points. So adenosine always meshes with thymine,3 and cytosine always meshes with guanine.
In our zipper analogy, any of these bases may happen to fall on either side of the zipper. So when the slider—represented by a polymerase enzyme—comes along, it can only join an A from one side with a T from the other, or a C with a G. At first this would seem to create a simple binary code: “A-or-C, A-or-C, A-or-C,” but the situation is more complex, because one side of the zipper can have any of the four bases in any order at each position. So the choice is actually “A-C-G-or-T, A-C-G-or-T, A-C-G-or-T.” This makes for a much richer information value, because the code now has four letters in any order, instead of the one from our simple mechanical zipper.
But this complexity also makes for a much more complex matching process as the two separate strands come together to complete the DNA molecule. The A in one strand might find a complementary T, or the C find a G, but if the next letter in line does not represent its opposite partner—if the sequence doesn’t match—then the zipper will buckle and jam.
|
Mostly, this is not a problem, because DNA usually doesn’t zip like our modern clothing fastener. Instead, when DNA gets copied in the nucleus just before the cell divides, the two conjoined strands slip apart, or unzip, in a process called “denaturing.” Then the polymerase enzyme simply assembles a complement—or reverse letter coding—for each single strand from among a sea of loose bases, rather like matching up the buttons in a sewing kit. Or again, when the DNA unwinds and gets transcribed into messenger RNA, that complementary strand is assembled from loose bases that are selected to match the next letter in line.
For a long time, molecular biologists believed that DNA existed only to be transcribed into messenger RNA, which was then translated into proteins out in the cell body. This was the “central dogma” of genetics. According to this teaching, DNA’s only purpose was to create messenger RNA—and also to replicate itself accurately during cell division, so that each daughter cell in a growing organism got a correct copy of the code.
After researchers had finished sequencing the human genome and spelled out every letter of the code—this was back around the year 2000—they discovered that less than 10% of the three billion base pairs of human DNA were used for coding proteins. But they still clung to the dogma. They ruled that the other 90% had to be “junk,” or old coding left over from our genetic ancestors, and was no use to anyone now.4 But within a couple of years, with more study of cellular processes, genetic researchers began to detect short, single strands of RNA only about fifty or a hundred base pairs long. These tiny strands, called “microRNAs,” were unlike messenger RNA in that they didn’t seem to leave the cell’s nucleus. Instead, they stayed inside and seemed to be involved in a process called “gene silencing” or “RNA interference.”
Human thinking quickly evolved to see that these strands of microRNA are the main way the cell differentiates itself during embryonic growth and development. That “other 90%” of the nuclear DNA serves to produce these microRNAs, which float around inside the nucleus and settle on complementary strands of DNA—in a process called “annealing”—to promote or inhibit a gene’s production of its messenger RNA. If you think of the 10% of DNA which represents the protein-coding genes as the body’s parts list, then the 90% of DNA which produces microRNAs is the body’s instruction set and assembly manual.
Amazingly, complementary strands—where every A meets a T, and every C meets a G—can find and mesh themselves over long strings of letters that happen to lie far apart in the code. The covalent bonds align with each other evenly, usually without buckling or breaking.5 This process of annealing a fragment of microRNA to its corresponding nuclear DNA is at least one case where an existing code string must find its exact complement—an A for each T, a C for each G, letter perfect all down the line. If a string of fifty or more bases tried to anneal to a complementary strand that had even just one or two letters out of place, the strand would buckle and jam, like a broken zipper.6
It’s an amazing feat of chemistry that draws these two strands of complementarily bonding molecules together over relatively long distances within the tangle that is the usual state of a free-floating DNA molecule. It’s even more amazing that they can orient themselves and match up perfectly, like the two halves of a zipper just happening to wrap around and snug their teeth together without the benefit of a mechanical slider. You might even call it a miracle—if you believed in that kind of thing.
1. Some of the newer models have other configurations, like grooves and ridges. Same principle.
2. Another pyrimidine base—uracil, or U—substitutes for thymine when the DNA strand is transcribed into its complementary RNA strand. Why? Well, it’s thought that DNA is actually a later evolutionary advancement on RNA. After all, ribose nucleic acid—with an OH group attached to the second carbon in the ring—had to lose that oxygen atom in order to become deoxy ribose. And adding a methyl group (CH3) to uracil turns it into thymine. In both cases—losing the oxygen and adding the methyl—increases the stability of the DNA molecule. Since the purpose of DNA is to preserve a coding system over a long period of time, stability is an evolutionary goal.
On the other hand, RNA serves a relatively ephemeral purpose in the genetic system. It carries the code from the DNA molecule in the nucleus to the protein-making machinery out in the cell body, where the code coordinates the stringing together of amino acids into a long-chain protein sequence. In fact, it’s probably better if RNA strands degraded quickly; otherwise they might hang around and get used to make second and third copies of the protein and so disrupt the cell’s functions.
3. Or uracil again.
4. But one of my colleagues at the genetic analysis company disputed this notion early on. Copying DNA takes a lot of energy, she said, because of that phosphate bond in the DNA molecule’s backbone. The phosphate bonds of the molecule adenosine triphosphate, or ATP, are the source of the cell’s energy. These bonds are created in the mitochondria from the chemical energy in our food and released as ATP into the cell body. Different cellular processes then break these bonds in order to drive chemical reactions. It made no sense to my colleague for the cell to spend all that energy in the replication of junk DNA. So, she reasoned, that other 90% of the genome had to have a purpose.
5. Although sometimes the matchup can get confused if the sequence has long strings of identical letters, like A-A-A-A-A-A-A.
6. Genetic analysis makes use of this strand-to-strand annealing capability. By creating the complementary strand to a known DNA sequence, we can find and latch onto a random sample of DNA and amplify it in the process of polymerase chain reaction, or PCR. This amplification has many uses by determining the sequence of coding beyond the annealing patch in a DNA strand—from identifying individuals in paternity and forensics cases to identifying different mutations to a known gene.