
 |
|
|
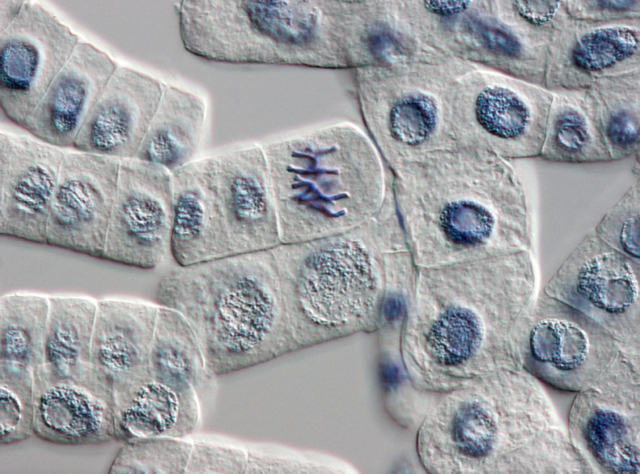
Onion cells dividing |
When I joined Applied Biosystems in the spring of 2000, the Human Genome Project Centers and our sister company Celera were just finishing up and publishing the first draft of the human genome, which had been made possible by our own high-volume sequencing machines. One of the surprises from this first draft was that only about ten percent of the human body’s DNA is involved in coding for proteins—and protein-coding had become the offhand definition of the word “gene.”1 The other ninety percent of the genome was a mystery. Many people at the time began calling this ninety percent “junk DNA.” By this they meant leftovers from our species’ long evolution, old genes we no longer used and which were slowly degrading by random mutation into genetic mush.
One day, however, I was walking across campus with one of our chemists who doubted the junk hypothesis. The human body, she said, invests way too much energy in copying the DNA of every cell every time it divides. Why would we waste that energy copying junk? This is obvious, of course, when you remember that the same phosphate bonds used in the backbone of the DNA molecule’s single strands are also used in the adenosine triphosphate molecule, which supplies energy throughout the cell. Phosphorus is a relatively rare element in the body. You don’t want to go sequestering it inside junk strands.
A year or two later, genetic scientists began focusing on short strands of transcribed DNA that were only about twenty to fifty base pairs long, which they dubbed “microRNAs.” These bits of material were first associated with “gene silencing.” Researchers had observed that adding them to the cells of a plant, say, could turn a variety with the genes to produce a red flower into one that produced a white flower—silencing the genes for red pigment.
It wasn’t long after this, about 2004, that Applied Biosystems hosted an in-house lecture by Eric Davidson, a researcher at the California Institute of Technology. He was working with sea urchin embryos and had discovered an intricate dance that occurs in the nucleus of the cells in a blastula—the hollow sphere that forms right after the egg is fertilized and right before the embryonic cells began differentiating into the sea urchin’s various body parts. By sacrificing tens of thousands of sea urchin eggs at spaced intervals following fertilization and studying their nuclear DNA, he was able to trace a pattern of microRNAs being transcribed from the DNA, annealing to other patches of the DNA inside the nucleus, and promoting the transcription of even more microRNAs. This cascading pattern differed among the cells, depending upon where in the blastula they resided and how much time had passed since fertilization. Davidson traced out this cascade of microRNAs, compared it to the cascades found in species that had not shared a common ancestor with sea urchins for many millions of years, and determined that this cascade of differentiation was highly conserved. That is, it’s probably common to all multi-celled life, including our own.2
Eric Davidson’s work suggests that, while ten percent of the genome codes for the messenger RNAs that go out into the cell body and become translated by the ribosomes into proteins—in other words, this is our body’s parts list—the other ninety percent codes for all these tiny microRNAs that stay inside the nucleus and control the timing of a cell’s development and differentiation into various tissues, organs, and body parts. That is, this ninety percent of DNA constitutes our body’s assembly manual.
It was three or four years later, while I was still at Applied Biosystems, that scientists began discussing a new field called “epigenomics.”3 In its most common form, methyl groups (CH3) become attached to one or more C-G sequences in the promoter regions that lie just ahead of protein-coding gene sequences. Where this occurs, the transcription factors that normally bind to the region and force the gene’s transcription into RNA are blocked, and the gene is silenced or suppressed—that is, it cannot be selected for expression and is no longer used by that cell to make proteins.4
Davidson showed how cells became differentiated by selectively promoting the expression of various genes. Liver cells, bone cells, brain cells, tooth-budding cells—all become what they are in the final adult organism by differentially expressing the genes that are available in the complete copy of the genome that’s inside every cell. But how does the body stop that expression? Some proteins are only needed by the cell when the embryo is developing, such as when limbs are budded and grow outward from the embryo’s developing trunk, or when fingers and toes are multiplied across the ends of the sprouting arms and legs. Some proteins are only needed at certain juvenile stages, such as when we grow our first set of baby teeth, or again when we grow our adult teeth at around age seven. Then cells in the jaw begin secreting the bonelike dentine and the hardened mineral called enamel to create a tooth. But after the tooth is formed, they must stop secreting these materials.
If some method of shutting off the genes did not exist, chaos would surely follow. Every cell in the body would try to reproduce every protein the body ever needed, all the time, clogging and rupturing the cell. Or cells might randomly express proteins not associated with their function: brain cells producing liver proteins, liver cells producing brain proteins, skin cells producing patches of enamel, until the body fell apart.
Methylation and the other ways of shutting off a gene appear to be the solution to this problem. Once a cell has achieved its primary function, the chemical addition of methylation shuts down its development and enables only the proteins needed in its adult life. And we know this must be so because a distinct enzyme, methyltransferase, exists to copy the methylation pattern of the adult cell’s DNA whenever the cell divides, so that the daughter cells each inherit the original’s suppression configuration.5
The epigenome is usually spoken of as some kind of environmental accident. A gene’s promoter region is said to become methylated—presumably because of too many methyl groups floating around in the environment—and it unpredictably shuts down the gene. This action of the epigenome is usually cited for the differences found in identical twins as they mature. Although pairs of young twins may be virtually indistinguishable as babies, toddlers, and children, older twins—especially those raised apart, in different environments—may appear quite different, even though both have the same gene set.
I would maintain that this kind of accidental variability is a side effect of the epigenome’s operation. The main show is stopping the promotion of genes after their developmental usefulness has passed and so settling the cell as a single tissue type. The existence and function of methyltransferase tends to suggest this.
The epigenome and the selective methylation of genetic promoter regions is one of the most important parts of cell growth, development, and function. And up until the last ten years, scientists had no notion of its existence. Like microRNAs—those tiny bits of transcribed DNA that never go on to code for proteins—the epigenome was an undiscovered country, a function and the solution to a problem that geneticists had never even considered, until we began probing the genome and the mystery of how the four-base code becomes a microbe, a plant, an animal, or a human being.
Research into the origins and chemical nature of life accelerated a hundredfold after the deciphering of, first, the human genome and, then, the genomes of many other organisms which are used for study or as stand-ins for human biology. Now epigenetic studies, as well as the study of microRNAs and other aspects of the genetic code, are proceeding apace at universities, institutes, and industrial laboratories all over the world. The results of these research efforts are shared almost hourly by electronic and print journals. Molecular biologists are closing in on a complete understanding of the human body, of the cell, and all its systems—not just in genomics or epigenomics but in other areas like proteomics (the study of proteins), metabolomics (the study of metabolic inputs and outputs), biomics (the study of the body’s microbial ecology), and all the other “–omics” yet to come.
It is my belief that in twenty years or so, plus or minus a decade, we will have a pretty good picture of the entire chemical nature of life itself. … And then the fun will begin.
1. That’s according to the old “central dogma” of molecular biology, which stated that the flow of information was always from the nuclear DNA, which was transcribed into messenger RNA, which then migrated out into the cell body, where it was translated into proteins. As this article shows, we’ve learned a lot since then.
2. See also the stories told in The Flowering of Life from August 25, 2013 and “Gene Deserts” and the Future of Medicine from December 5, 2010.
3. “Epigenome” refers to chemical changes to the genome, the DNA inside the cell’s nucleus, and to the arrangement of its strands around protein blocks called histones around which the chromosomes coil. Nothing in the epigenome changes the base-pair sequences of the genome itself, merely the body’s access to them in making proteins. The word derives from the Latin and Greek root epi, meaning “upon,” “on,” or “to.”
4. This is the most common pattern: methylation in the promoter area to suppress a gene. But in some cases methylation within the coding sequence itself has been shown to stop expression. And now researchers are also learning that enhancer regions—patches of DNA not adjacent to a gene itself, like the promoter, and sometimes quite far away from it—can become methylated. When this happens, the gene may be variably suppressed. That is, it’s not just turned on or off, but sometimes on and sometimes off, depending on conditions that are yet to be understood. Suppression of enhancer regions seems to be more often associated with genes involved in cancer. See DNA Methylation in Cancer Goes the Distance via Enhancers.
Another method of gene regulation is histone modification, which changes the protein blocks around which the DNA strands of a chromosome are wound. In one case, an enzyme adds an acetyl group (CH3CO–) to the histone, changing its positive electrical charge and so reducing its attraction for negatively charged DNA. This loosens the coiled DNA strand, so that it can be more easily transcribed and the genes in that area activated. In other cases case, the arrangement of the chromatin fiber itself—the sequential pieces of DNA as they wrap around nearby histone proteins—is remodeled by one of several chemical changes, such as methylation, acetylation, phosphorylation (addition of a phosphate group, PO43–), ubiquitination (addition of a small regulatory protein found in almost all tissues—that is, “ubiquitously”—of multi-celled organisms), and other chemicals.
5. Methylation and the other chemical methods of epigenomics also seem to be the key to inducing pluripotency—that is, capability of becoming more than one cell type—in creating stem cells from the normal, tissue-typed cells in the body. If you can strip away some or all of the methyls and other groups that lock a cell into functioning as one kind of tissue or another, you can make it more adaptable and available for reprogramming.